Drop a spec of E. coli bacteria into a cup of sugar water and overnight that clear liquid will transform into a cloudy soup containing a hundred billion (1011) living cells.

Now, into your cup of E. coli bacteria, add a touch of RNA bacteriophage—called “phage” for short—-and in a few hours the cloudy soup will turn clear again. The once thriving E. coli?…dead. In their place, invisible to the naked eye, are a hundred trillion (1014) new phage.

RNA phage are viruses that infect bacteria. Like other RNA viruses, they’re extreme minimalists, consisting of a protein shell called a “capsid” surrounding an RNA genome encoding only a few genes. The phage we work with in our lab, named MS2, is made up of 180 molecules: 178 copies of its coat protein, one copy of its maturation protein, and one copy of its genomic RNA. That’s it! Armed only with these two-hundred-or-so molecules, viruses like MS2 are able to infect their hosts, highjack the host cells’ machinery, and replicate in tremendous yield.
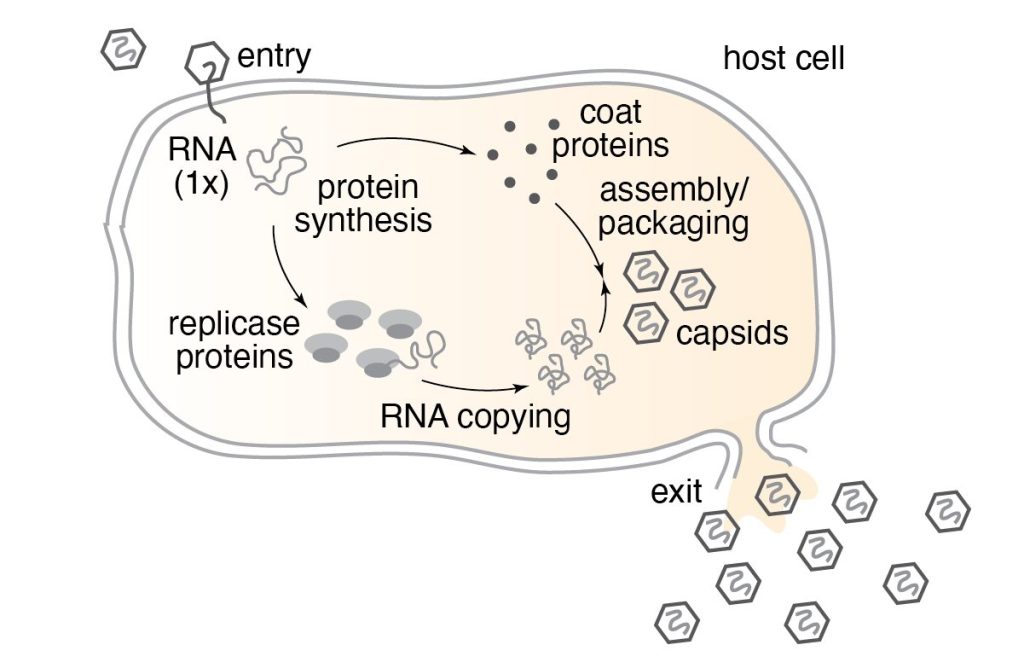
Because they’re made from so few molecules, RNA viruses can be studied and understood “from the bottom up” using physical chemistry. In our lab, we take the viruses apart, we separate their protein and RNA, and then we put them back together. We measure the structure and dynamics of the viral molecules in isolation, and then we measure collective properties that emerge when the molecules interact with eachother. We team up with theorists and computational chemists to turn our measurements into quantitative models that help us understand how viruses perform complex biological functions, like infection.
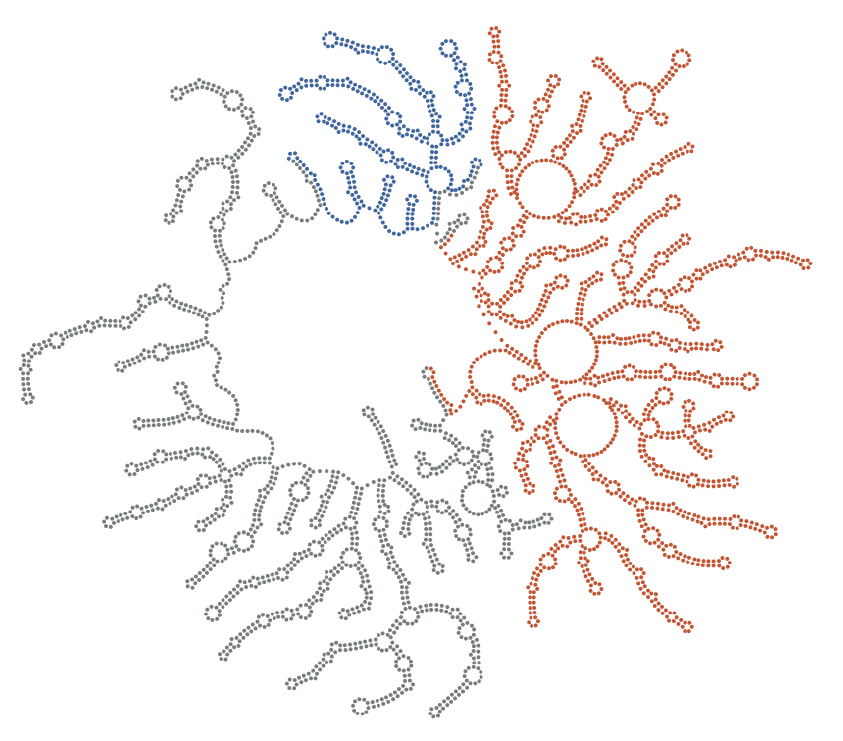
We are especially fascinated by the process of “self-assembly”, which is when an ordered structure appears spontaneously from an initially disordered mixture of molecules. RNA viruses are experts in self-assembly—it’s how they build their protein capsids and fold their RNA genomes—and one cannot fully appreciate nor understand how these viruses operate without understanding how they self-assemble. Our lab uses ultra sensitive optical microscopes and tools from a field called “DNA nanotechnology” to measure viral self-assembly at small length scales (nanometers) and over a broad range of time scales (milliseconds to days).
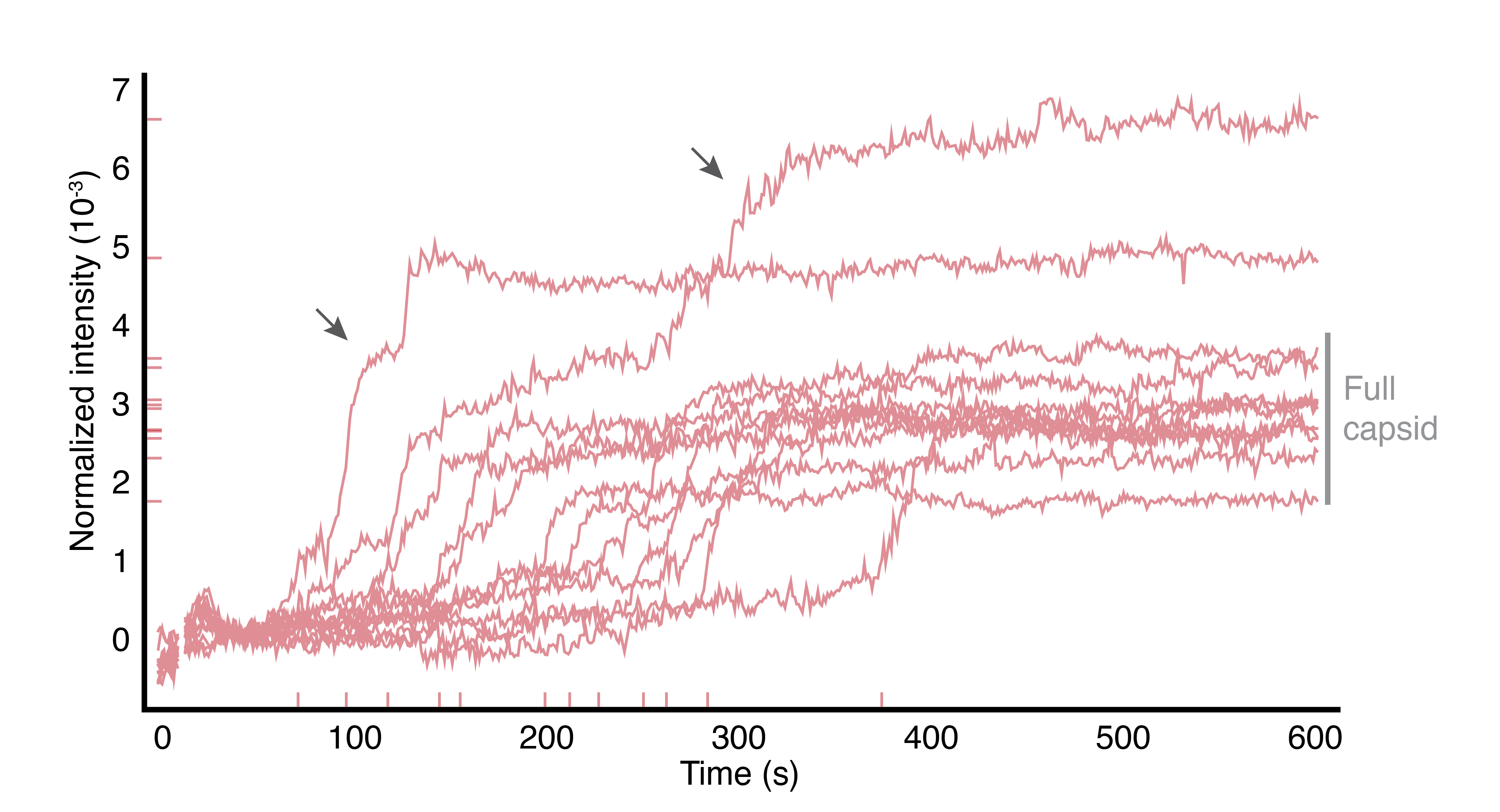
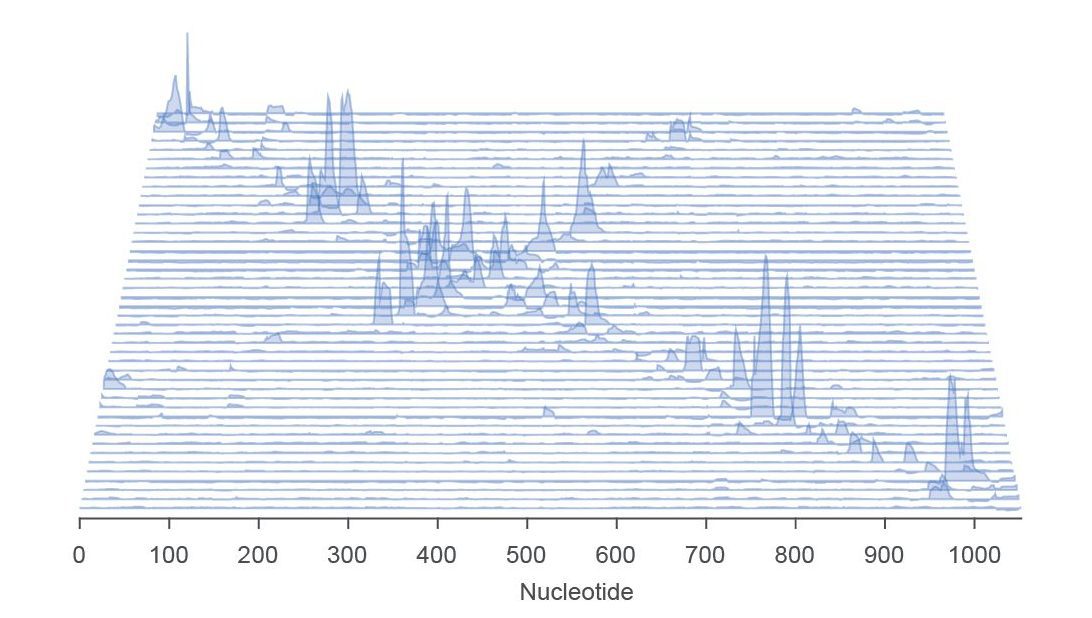
In addition to teaching us about how viruses work, we hope our results will inspire new ways of repurposing viruses to do useful things, such as delivering therapeutic cargos to cells. And we also hope to uncover new ways to block the spread of harmful viruses that cause disease.